Codons do not specify amino acids!
Codons specify tRNA molecules. tRNA molecules specify amino acids. Let’s take a look at some of the features of a tRNA molecule and imagine that we are an amino acid, Leucine, for instance, encountering our very own tRNA.
Each tRNA molecule is a polynucleotide, as are mRNA and DNA, and each tRNA folds into a three-dimensional shape that determines its function, much like a protein. This does not stop tRNA from being and behaving like DNA and mRNA with respect to nucleotide-nucleotide interaction. Each tRNA has an anticodon region which gives the molecule its raison d'être - complimentary pairing with codons. However, there are other nucleotides in the molecule that play a key role in the translation process, many of which can be schematically found in the “variable region” or body as labeled above.
This is a very stylized look at tRNA, but it will help us make the central point about the role of tRNA in the genetic code. The actual structure of tRNA is fairly well documented, but this simplified view is copasetic with what is known. We can revise our assignment table, substituting the tRNA-amino acid complex where we previously only had amino acids.
Nothing seems to have fundamentally changed. This is because we have not done enough thinking about our new partner, tRNA. A much better description can be found elsewhere on the internet, and here is a link: http://www.mun.ca/biochem/courses/3107/Lectures/Topics/tRNA.html
Here is some interesting information about tRNA that can be found at that link:
In 1966, Francis Crick proposed the Wobble hypothesis… He suggested that while the interaction between the codon in the mRNA and the anticodon in the tRNA needed to be exact in two of the three nucleotide positions, this did not have to be so in the third position. He proposed that non-standard base-pairing might occur between the nucleotide base in the 5' position of the anticodon and the 3' position of the codon.This hypothesis not only accounts for the number of tRNAs that are observed, it also accounts for the degeneracy that is observed in the Genetic Code. The degenerate base is that in the wobble positionThese wobble rules are not followed exactly. If they were, only 31 tRNAs would be needed to pair will all possible codons. There are, however, more than 31 tRNAs.The sequence of Escherichia coli K12 contains 84 tRNA reading frames. The only amino acids with a single tRNA are histidine, tryptophan and selenocysteine. There are 7 tRNAs for arginine and valine, and there are 8 tRNAs for leucine.
Let’s incorporate this newfound info into our assignment table. Since we don’t have the specifics on an organism’s distribution of tRNA, again we will wing it in a stylized way. This will help us visualize how a hypothetical population of tRNA, say E. Coli, might be distributed across the genetic code.
The above discription of the population and wobble appears to me to be a direct contradiction. The wobble hypothesis was presented to explain redundancy, not create it. Furthermore, I can’t possibly imagine what definition of linear this concept might fit into. Cause and effect are now clearly disproportionate. Our conventional view of a one-codon-one-amino acid system can at least be plotted on a graph, and it actually produces a line.
But with this new realization that codons do not specify amino acids, they specify tRNAs we must revisit the graph. If we plot every tRNA against its codon, and there are more tRNAs than codons, then we cannot produce a line. Depending on the exact proportions, we might get a plot that will look more like an electron cloud.
There is a difference between reading frames and actual tRNA in a cell. But we also haven’t gotten all the potential forms of transcriptional splicing and post-transcriptional modifications of tRNA into the mix. The bottom line is that we presently do not know the precise population of discrete tRNA molecules in any given cell. There could be less than expected, or there could be more, a lot more. This will become an important part of sorting out the actual genetic code.
We now have a problem when we assemble proteins on the shop floor. Before in the linear model, when an order for an amino acid came down from nucleotides, all we had to do was refer to our linear graph and assemble the amino acid - the only amino acid - into the chain as dictated by the code. We now must find a way to decide which of these damn tRNA to use. If two different tRNA will work for a single codon, how do we decide which one to use? We pine for the simpler world of yesteryear. Why do we need to confuse things, why not stick to a one-to-one system?
If wobble is a beneficial phenomenon credited with reducing redundancy, then it should actually in some way reduce redundancy. A force that allows a property of a system to exceed its predicted maximum should never be proposed as a reducing force. Codons are now ambiguous with respect to the tRNA they specify, and there is a likelihood of finding more than 64 tRNA in a cell.
Strictly from the standpoint of information, wobble rules alone increase the information content of translation. The rules allow for an expansion of molecular choices at the third position.
Wobble is touted as an information reducer, but in fact it increases the information by 250% in converting 64 codons into 160 anti-codons. Consequently, the anticodon table is more robust than the codon table.
Translation mechanisms to handle the newfound ambiguity can be leveraged to increase the information even further. In other words, anticodons are just the start. I am uncertain of whether there is an upper boundary to the number of distinct tRNA that might be found in the cell. Reliable sources tell me there are ~1000 known reading frames for tRNA so far located in the human genome. Who knows what this translates into in terms of recombination and post-transcriptional modifications. The actual number of human tRNA might be surprisingly large.
We must try a little harder to come up with a model that actually explains these numeric contradictions with dogmatic expectations, rather than ignores them. Take Leucine, for instance. We know that Leucine is highly redundant with respect to codon assignments (it has six). The wobble hypothesis as a reducer predicts that Leucine might only require two tRNA molecules to be fully translated in the code. This is because all Leucine codons start with one of two diads: UU and CU.
Leucine codons: UUA UUG CUU CUC CUG CUA
So leucine would only need anti-codons of AAI and GAI. However, now we are told that Leucine has perhaps eight tRNA molecules in E. Coli, which makes little sense. It makes even less sense when one considers that without wobble one might expect six (one for each codon) and the wobble hypothesis was ostensibly proposed to reduce this number even further. There is now ambiguity in the translation of at least one codon.
Every tRNA molecule carries an identical Leucine, but there are differences in the nucleotide quantities and sequences of each tRNA. These differences are schematically demonstrated above by changing the colors of the anticodons at the bottom of the molecule and the variable regions in the middle of the molecules.
I do not have specifics of the actual variations or the pattern made by the group, but we can guess that at least two sets of anticodons probably share sequences. We also know that this phenomenon grows much larger in eukaryotes than in the prokaryote example of E. Coli that we are addressing. Let’s put this concept visually into the assignment table.
Note that these molecules are fairly large compared to the size of a codon. Each one has between 73 and 93 nucleotides, which means that the anticodon region accounts for 4% of the molecule at most. The amino acid is truly tiny compared to the tRNA molecule. The whole function of these molecules is to bring these tiny amino acids into proximity so they can be bonded together in a polypeptide chain. The steric influence of these molecules must be significant. To get a better view of this I will show a stylized version of translation of four amino acids using these tRNA. We will stick with Leucine for this illustration, but we will use a different tRNA for the addition of each residue.
This would be a truly remarkable feat of molecular manners to find anything approaching this type of well-behaved stereochemistry in nature. For starters, it is unlikely that all four tRNA leucines will ever simultaneously line-up on mRNA as pictured, they generally work two at a time, but all four will play a role in the tetra-leucine at some time.
Think of the chain of tRNA as a virtual scaffolding that forms through time. A polypeptide with 100 residues will never visit a contemporaneous scaffold of 100 tRNA, but we can think of it that way. The scaffold is positively huge compared to the polypeptide for which it forms a template; this diagram does not do justice to the relative size differences. It is not implausible to think that the topology of the scaffold will impact the primary structure of a polypeptide. The question now becomes, how many different scaffolds can be built in a given cell that create the same amino acid sequence? How do these discrete scaffolds differ, and how will the different scaffolds affect the ultimate shape of the proteins they create?
The schematic demonstrates another critical point: the genetic code is operating in at least two distinct zones of interaction. There is nucleotide interaction obviously occurring between codons and anticodons, but there also is nucleotide interaction occurring between nucleotides in adjacent tRNA molecules. More significantly, there is a necessary spatial interaction between adjacent tRNA. It is this dual level of interaction that makes the code truly multi-dimensional, going beyond sequence. It also is this extra dimension of information, or interaction that translates the stereochemical information in the genetic code.
A revised view of this situation would lead to the conclusion that eight does in fact represent a dramatically reduced tRNA count for Leucine, due in large part to wobble. There could potentially be many more tRNA in a cell, and each one might contribute to a tRNA scaffold in a topologically different way.
tRNAs now compete for space, or jockey for position. There must be logical pairings between tRNA in order for a peptide bond to be made successfully. Consequently, the tiny amino acid and its peptide bond will be jostled, or sterically affected in the process. Small alterations in the properties of tRNA molecules could have large stereochemical consequences for translation. The tRNA act as angle compounding dies in the expansion of spatial information. They are geometrically suited – perfectly – for this task.
The common thread, or the encryption key involves the golden triangle. The shape first shows up in the cross-sectional decagon of the double helix.
This is the seed of spatial information that begins in nucleotides and is unpacked into proteins. The shape of tRNA preserve these logical proportions.
More fun, if we place a golden triangle solid spike on a dodecahedron to represent each anti-codon on the Rafiki map, we find an impressive fit for an expanded icosidodecahedron.
I find it absolutely remarkable that any regular shape can be found to symbolically represent tRNA molecules on a comprehensive mapping of all codons. It is one thing to realize that the real symbolic constraints of codons are an exact match with the symmetry elements of a dodecahedron, but it is another thing to discover that tRNA shapes will extend the symmetry much further. It gives us pause, during which we should consider the question: Did codons make this shape, or did this shape make codons?
If we let our imaginations flex their considerable muscles, we can see a clever, self-contained erector set for molecules, a platonic building system of blueprints, building blocks and angle compounding dies. From an engineering standpoint, it is perplexing that tRNA molecules should be so large while codons and amino acids are so small. When the first inkling of the existence of a functional molecule like tRNA was imagined, they were given the name ‘adapters’. It was guessed that these molecules were mirrors of codons, and that they were only three nucleotides big. Although it turns out that tRNA seem to be inordinately large for this role, their actual size in and of itself is proof of nothing. However, it is irresistibly fun to speculate about collections of really large golden triangles mingling with sets of much smaller golden triangles in some cryptic engineering project, optimized for precision alignment of tiny tetrahedrons. It would be fabulous if they actually turn out to be the spatial amplifiers that they pretend to be.
Beyond this level of translation, downstream into folded proteins, structures are highly conserved in a surprisingly small number of shapes. The laws of form are precisely what drove D’Arcy Thompson at the turn of the century, and it has since been claimed that these conserved protein structures can be broadly classified by the symmetries they share with regular solids. Proteins love to form regular solids when they auto-assemble in larger numbers, so again we might find the golden mean at this level. It is not inconceivable then that the golden mean is the shared element through these various levels of genetic translation. At least it could be used as an intellectual tool to analyze information as it gets passed along and expanded in growing, organic systems. Self-assembly at the smaller scales, such as proteins and virus particles is a platonic game, but even larger structures, such as radiolarian shells, have long been known to have a fondness for platonic shapes.
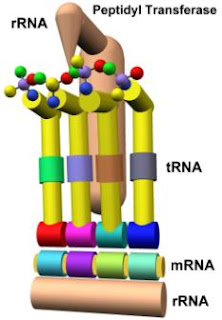
Returning to and further complicating the study of translation mechanisms, there is a third level of nucleotide involvement in the peptide bond formation. The catalyst for the bond is not a protein but a nucleotide, the 23S rRNA subunit. The catalytic property of this structure is referred to as peptidyl transferase, and it interacts with tRNA during bond formation and has a direct impact on the final state of the bond between every adjacent amino acid in every polypeptide.
Still another level of interaction is required. The tRNA must be ‘loaded’ with amino acids. This job is performed by a set of proteins, or enzymes known as transferases. There are twenty (one for each amino acid) and they are evenly divided into two types. The lines of division are clearly organized around proline.
Many interpretations of this distribution of two enzyme types have been made, but within the context of the Rafiki map the symmetry around proline is compelling.
More interesting, every tRNA accepts its amino acid onto the same three nucleotides, CCA, so as far as amino acids are concerned there is really only one codon. There is a separate code that operates in the world of transferases to identify anti-codons, amino acids, and something else.Perhaps these musings – thought experiments - do not prove a thing, but they do bring to light the pragmatic difficulties of taking a contrary position. There appear to be a huge number of nucleotides intervening between the codon and the amino acid, all of which seem to owe their invitation to the process to the need for a peptide bond. The consistency from this large and diverse collection of nucleotides in the process of making these peptide bonds has been proven, but uniformity of bonds has not. Therefore, the mechanism would seem to consistently form non-uniform bonds. Rafiki says that this bond irregularity is a pre-determined part of the program. It is the essence of genetic information. Nucleotides speak a stereochemical language in building the backbones of polypeptides.
In order for this view to be completely invalid, the following circumstances would have to be proven. For every potential sequence of our hypothetical tetra-leucine, identical tetra-peptides would have to be produced. Otherwise, the difference between tetra-peptides must be attributed to some macromolecular nucleotide mechanism acting on peptide bond formation. If we start with the assumption that there are 8 forms of tRNA-leucine available in a cell, then there are perhaps a minimum of 4096 code sequences that might specify tetra-leucine (84). Perhaps some tRNA-leucines are not sterically compatible with one or more of their brother tRNA molecules. In this case, when one tRNA follows an incompatible tRNA in the sequence the translation apparatus has a tRNA option (or perhaps a requirement). When this option does not exist, translation is terminated. There are tremendously more tRNA required by this system, and wobble can play an effective role in keeping this number smaller than it might otherwise have been.
We can now fully appreciate what it means to be an amino acid. We are leucine, after all – remember? The thought experiment asks one question, what do we see, as amino acids, at the point of contact with the genetic code. This is, in fact, what the genetic code is. It is all about nucleic acids talking to amino acids and getting them to behave in a way that leaves no doubt that information is crossing molecular boundaries. The money shot of this whole thread, so to speak, is in trying to decide what the nucleic acids are telling the amino acids. We now have two basic choices and we are going to use tetra-leucine to decide which one is more logical. If all conceivable nucleic acid sequences for four consecutive leucines “look” the same, or homogeneous to us as leucine, then the code probably can carry no stereochemical information.
However, if any valid scaffold has a different look to us, one that results in even a slightly different polypeptide backbone, and this structural difference leads to discrete folding options, then we must conclude that the code is truly multi-dimensional and it carries stereochemical information. A different “look” to the above sequence could take on any one of roughly 4000 appearances to us, assuming that we are leucine in E. Coli as described.
This is the view of a simple case, tetra-leucine. What is the view of our hypothetical 100 residue backbone? How many different topologies can the tRNA scaffolds generate? How do these differences translate into the information content of proteins? These are all good questions that to my eye haven’t even been asked, let alone answered. This institutional absence of curiosity is ludicrous, because after all, this is the genetic code. Dogma be damned.
We should realize that every organism can and does have a different complement of tRNA. From this standpoint, the assumption that all peptide bonds emerge from translation in a perfectly identical conformation seems untenable. If translation is causing this lack of homogeneity in peptide bonds then we must identify the information behind this, wherever it may reside.
The first significant impact of this realization is that we can identify the location of the genetic code. The genetic code resides in RNA. The process of translating nucleic acids into amino acids is an entirely RNA phenomenon. It involves many molecules of RNA, not just mRNA, and all involved RNA molecules must be viewed as a part of the genetic code. It is a stereochemical phenomenon, not one-dimensional sequence-only, and it contains stereochemical information. Codons reside on mRNA. They specify tRNA, but each tRNA interacts with every other tRNA. It is the tRNA-tRNA interaction in a sequence that defines every peptide bond in a protein. Finally, rRNA interacts with both mRNA and tRNA to complete the apparatus that builds each and every peptide bond. It is this complement of RNA – mRNA, tRNA, and rRNA – that defines the genetic code of protein synthesis for each organism.
The second significant impact of this thinking is the conclusion that the genetic code is far from universal. Not even the assignment table is universal, but this is just the tip of the non-universal iceberg. There is potentially a universal code, or code limit, but no known organism would need or ever approach this limit. If tRNA is an integral part of the code, and tRNA varies widely between organisms, then the code varies widely between organisms as well. We can visualize this with the following pair of schematics.
This is a return to the visualization of codons matching with the tetrahedrons of amino acids. This is not complete, and therefore it is not an accurate representation of “the genetic code”. This is a relatively complete representation of the correlation table, or assignment table of codons to amino acids, but it ignores many known features of the code. This is a codon table, and codons are all about mRNA. To this we now must add visualization for a network of tRNA.
This is a more accurate picture of what a genetic code might ‘look’ like in a single organism. Change a single nucleic acid in a single tRNA and you have probably changed the code in that organism. This is a truly networked view of the genetic code. It is a dynamic network of RNA.
The really fun thing is that we can now standardize, objectify, and maximally compress this view. That is the view of the Rafiki map. The map itself does not illustrate specific tRNA populations, but it provides the most logical foundation into which they can be placed. The relative positions of all the code’s components can now take shape.
Each tRNA molecule is a polynucleotide, as are mRNA and DNA, and each tRNA folds into a three-dimensional shape that determines its function, much like a protein. This does not stop tRNA from being and behaving like DNA and mRNA with respect to nucleotide-nucleotide interaction. Each tRNA has an anticodon region which gives the molecule its raison d'être - complimentary pairing with codons. However, there are other nucleotides in the molecule that play a key role in the translation process, many of which can be schematically found in the “variable region” or body as labeled above.
This is a very stylized look at tRNA, but it will help us make the central point about the role of tRNA in the genetic code. The actual structure of tRNA is fairly well documented, but this simplified view is copasetic with what is known. We can revise our assignment table, substituting the tRNA-amino acid complex where we previously only had amino acids.
Nothing seems to have fundamentally changed. This is because we have not done enough thinking about our new partner, tRNA. A much better description can be found elsewhere on the internet, and here is a link: http://www.mun.ca/biochem/courses/3107/Lectures/Topics/tRNA.html
Here is some interesting information about tRNA that can be found at that link:
In 1966, Francis Crick proposed the Wobble hypothesis… He suggested that while the interaction between the codon in the mRNA and the anticodon in the tRNA needed to be exact in two of the three nucleotide positions, this did not have to be so in the third position. He proposed that non-standard base-pairing might occur between the nucleotide base in the 5' position of the anticodon and the 3' position of the codon.This hypothesis not only accounts for the number of tRNAs that are observed, it also accounts for the degeneracy that is observed in the Genetic Code. The degenerate base is that in the wobble positionThese wobble rules are not followed exactly. If they were, only 31 tRNAs would be needed to pair will all possible codons. There are, however, more than 31 tRNAs.The sequence of Escherichia coli K12 contains 84 tRNA reading frames. The only amino acids with a single tRNA are histidine, tryptophan and selenocysteine. There are 7 tRNAs for arginine and valine, and there are 8 tRNAs for leucine.
Let’s incorporate this newfound info into our assignment table. Since we don’t have the specifics on an organism’s distribution of tRNA, again we will wing it in a stylized way. This will help us visualize how a hypothetical population of tRNA, say E. Coli, might be distributed across the genetic code.
The above discription of the population and wobble appears to me to be a direct contradiction. The wobble hypothesis was presented to explain redundancy, not create it. Furthermore, I can’t possibly imagine what definition of linear this concept might fit into. Cause and effect are now clearly disproportionate. Our conventional view of a one-codon-one-amino acid system can at least be plotted on a graph, and it actually produces a line.
But with this new realization that codons do not specify amino acids, they specify tRNAs we must revisit the graph. If we plot every tRNA against its codon, and there are more tRNAs than codons, then we cannot produce a line. Depending on the exact proportions, we might get a plot that will look more like an electron cloud.
There is a difference between reading frames and actual tRNA in a cell. But we also haven’t gotten all the potential forms of transcriptional splicing and post-transcriptional modifications of tRNA into the mix. The bottom line is that we presently do not know the precise population of discrete tRNA molecules in any given cell. There could be less than expected, or there could be more, a lot more. This will become an important part of sorting out the actual genetic code.
We now have a problem when we assemble proteins on the shop floor. Before in the linear model, when an order for an amino acid came down from nucleotides, all we had to do was refer to our linear graph and assemble the amino acid - the only amino acid - into the chain as dictated by the code. We now must find a way to decide which of these damn tRNA to use. If two different tRNA will work for a single codon, how do we decide which one to use? We pine for the simpler world of yesteryear. Why do we need to confuse things, why not stick to a one-to-one system?
If wobble is a beneficial phenomenon credited with reducing redundancy, then it should actually in some way reduce redundancy. A force that allows a property of a system to exceed its predicted maximum should never be proposed as a reducing force. Codons are now ambiguous with respect to the tRNA they specify, and there is a likelihood of finding more than 64 tRNA in a cell.
Strictly from the standpoint of information, wobble rules alone increase the information content of translation. The rules allow for an expansion of molecular choices at the third position.
Wobble is touted as an information reducer, but in fact it increases the information by 250% in converting 64 codons into 160 anti-codons. Consequently, the anticodon table is more robust than the codon table.

Translation mechanisms to handle the newfound ambiguity can be leveraged to increase the information even further. In other words, anticodons are just the start. I am uncertain of whether there is an upper boundary to the number of distinct tRNA that might be found in the cell. Reliable sources tell me there are ~1000 known reading frames for tRNA so far located in the human genome. Who knows what this translates into in terms of recombination and post-transcriptional modifications. The actual number of human tRNA might be surprisingly large.

We must try a little harder to come up with a model that actually explains these numeric contradictions with dogmatic expectations, rather than ignores them. Take Leucine, for instance. We know that Leucine is highly redundant with respect to codon assignments (it has six). The wobble hypothesis as a reducer predicts that Leucine might only require two tRNA molecules to be fully translated in the code. This is because all Leucine codons start with one of two diads: UU and CU.
Leucine codons: UUA UUG CUU CUC CUG CUA
So leucine would only need anti-codons of AAI and GAI. However, now we are told that Leucine has perhaps eight tRNA molecules in E. Coli, which makes little sense. It makes even less sense when one considers that without wobble one might expect six (one for each codon) and the wobble hypothesis was ostensibly proposed to reduce this number even further. There is now ambiguity in the translation of at least one codon.
Every tRNA molecule carries an identical Leucine, but there are differences in the nucleotide quantities and sequences of each tRNA. These differences are schematically demonstrated above by changing the colors of the anticodons at the bottom of the molecule and the variable regions in the middle of the molecules.
I do not have specifics of the actual variations or the pattern made by the group, but we can guess that at least two sets of anticodons probably share sequences. We also know that this phenomenon grows much larger in eukaryotes than in the prokaryote example of E. Coli that we are addressing. Let’s put this concept visually into the assignment table.
Note that these molecules are fairly large compared to the size of a codon. Each one has between 73 and 93 nucleotides, which means that the anticodon region accounts for 4% of the molecule at most. The amino acid is truly tiny compared to the tRNA molecule. The whole function of these molecules is to bring these tiny amino acids into proximity so they can be bonded together in a polypeptide chain. The steric influence of these molecules must be significant. To get a better view of this I will show a stylized version of translation of four amino acids using these tRNA. We will stick with Leucine for this illustration, but we will use a different tRNA for the addition of each residue.
This would be a truly remarkable feat of molecular manners to find anything approaching this type of well-behaved stereochemistry in nature. For starters, it is unlikely that all four tRNA leucines will ever simultaneously line-up on mRNA as pictured, they generally work two at a time, but all four will play a role in the tetra-leucine at some time.
Think of the chain of tRNA as a virtual scaffolding that forms through time. A polypeptide with 100 residues will never visit a contemporaneous scaffold of 100 tRNA, but we can think of it that way. The scaffold is positively huge compared to the polypeptide for which it forms a template; this diagram does not do justice to the relative size differences. It is not implausible to think that the topology of the scaffold will impact the primary structure of a polypeptide. The question now becomes, how many different scaffolds can be built in a given cell that create the same amino acid sequence? How do these discrete scaffolds differ, and how will the different scaffolds affect the ultimate shape of the proteins they create?
The schematic demonstrates another critical point: the genetic code is operating in at least two distinct zones of interaction. There is nucleotide interaction obviously occurring between codons and anticodons, but there also is nucleotide interaction occurring between nucleotides in adjacent tRNA molecules. More significantly, there is a necessary spatial interaction between adjacent tRNA. It is this dual level of interaction that makes the code truly multi-dimensional, going beyond sequence. It also is this extra dimension of information, or interaction that translates the stereochemical information in the genetic code.
A revised view of this situation would lead to the conclusion that eight does in fact represent a dramatically reduced tRNA count for Leucine, due in large part to wobble. There could potentially be many more tRNA in a cell, and each one might contribute to a tRNA scaffold in a topologically different way.
tRNAs now compete for space, or jockey for position. There must be logical pairings between tRNA in order for a peptide bond to be made successfully. Consequently, the tiny amino acid and its peptide bond will be jostled, or sterically affected in the process. Small alterations in the properties of tRNA molecules could have large stereochemical consequences for translation. The tRNA act as angle compounding dies in the expansion of spatial information. They are geometrically suited – perfectly – for this task.
The common thread, or the encryption key involves the golden triangle. The shape first shows up in the cross-sectional decagon of the double helix.
This is the seed of spatial information that begins in nucleotides and is unpacked into proteins. The shape of tRNA preserve these logical proportions.
More fun, if we place a golden triangle solid spike on a dodecahedron to represent each anti-codon on the Rafiki map, we find an impressive fit for an expanded icosidodecahedron.
I find it absolutely remarkable that any regular shape can be found to symbolically represent tRNA molecules on a comprehensive mapping of all codons. It is one thing to realize that the real symbolic constraints of codons are an exact match with the symmetry elements of a dodecahedron, but it is another thing to discover that tRNA shapes will extend the symmetry much further. It gives us pause, during which we should consider the question: Did codons make this shape, or did this shape make codons?
If we let our imaginations flex their considerable muscles, we can see a clever, self-contained erector set for molecules, a platonic building system of blueprints, building blocks and angle compounding dies. From an engineering standpoint, it is perplexing that tRNA molecules should be so large while codons and amino acids are so small. When the first inkling of the existence of a functional molecule like tRNA was imagined, they were given the name ‘adapters’. It was guessed that these molecules were mirrors of codons, and that they were only three nucleotides big. Although it turns out that tRNA seem to be inordinately large for this role, their actual size in and of itself is proof of nothing. However, it is irresistibly fun to speculate about collections of really large golden triangles mingling with sets of much smaller golden triangles in some cryptic engineering project, optimized for precision alignment of tiny tetrahedrons. It would be fabulous if they actually turn out to be the spatial amplifiers that they pretend to be.
Beyond this level of translation, downstream into folded proteins, structures are highly conserved in a surprisingly small number of shapes. The laws of form are precisely what drove D’Arcy Thompson at the turn of the century, and it has since been claimed that these conserved protein structures can be broadly classified by the symmetries they share with regular solids. Proteins love to form regular solids when they auto-assemble in larger numbers, so again we might find the golden mean at this level. It is not inconceivable then that the golden mean is the shared element through these various levels of genetic translation. At least it could be used as an intellectual tool to analyze information as it gets passed along and expanded in growing, organic systems. Self-assembly at the smaller scales, such as proteins and virus particles is a platonic game, but even larger structures, such as radiolarian shells, have long been known to have a fondness for platonic shapes.

Returning to and further complicating the study of translation mechanisms, there is a third level of nucleotide involvement in the peptide bond formation. The catalyst for the bond is not a protein but a nucleotide, the 23S rRNA subunit. The catalytic property of this structure is referred to as peptidyl transferase, and it interacts with tRNA during bond formation and has a direct impact on the final state of the bond between every adjacent amino acid in every polypeptide.
Still another level of interaction is required. The tRNA must be ‘loaded’ with amino acids. This job is performed by a set of proteins, or enzymes known as transferases. There are twenty (one for each amino acid) and they are evenly divided into two types. The lines of division are clearly organized around proline.
Many interpretations of this distribution of two enzyme types have been made, but within the context of the Rafiki map the symmetry around proline is compelling.
More interesting, every tRNA accepts its amino acid onto the same three nucleotides, CCA, so as far as amino acids are concerned there is really only one codon. There is a separate code that operates in the world of transferases to identify anti-codons, amino acids, and something else.Perhaps these musings – thought experiments - do not prove a thing, but they do bring to light the pragmatic difficulties of taking a contrary position. There appear to be a huge number of nucleotides intervening between the codon and the amino acid, all of which seem to owe their invitation to the process to the need for a peptide bond. The consistency from this large and diverse collection of nucleotides in the process of making these peptide bonds has been proven, but uniformity of bonds has not. Therefore, the mechanism would seem to consistently form non-uniform bonds. Rafiki says that this bond irregularity is a pre-determined part of the program. It is the essence of genetic information. Nucleotides speak a stereochemical language in building the backbones of polypeptides.
In order for this view to be completely invalid, the following circumstances would have to be proven. For every potential sequence of our hypothetical tetra-leucine, identical tetra-peptides would have to be produced. Otherwise, the difference between tetra-peptides must be attributed to some macromolecular nucleotide mechanism acting on peptide bond formation. If we start with the assumption that there are 8 forms of tRNA-leucine available in a cell, then there are perhaps a minimum of 4096 code sequences that might specify tetra-leucine (84). Perhaps some tRNA-leucines are not sterically compatible with one or more of their brother tRNA molecules. In this case, when one tRNA follows an incompatible tRNA in the sequence the translation apparatus has a tRNA option (or perhaps a requirement). When this option does not exist, translation is terminated. There are tremendously more tRNA required by this system, and wobble can play an effective role in keeping this number smaller than it might otherwise have been.
We can now fully appreciate what it means to be an amino acid. We are leucine, after all – remember? The thought experiment asks one question, what do we see, as amino acids, at the point of contact with the genetic code. This is, in fact, what the genetic code is. It is all about nucleic acids talking to amino acids and getting them to behave in a way that leaves no doubt that information is crossing molecular boundaries. The money shot of this whole thread, so to speak, is in trying to decide what the nucleic acids are telling the amino acids. We now have two basic choices and we are going to use tetra-leucine to decide which one is more logical. If all conceivable nucleic acid sequences for four consecutive leucines “look” the same, or homogeneous to us as leucine, then the code probably can carry no stereochemical information.
However, if any valid scaffold has a different look to us, one that results in even a slightly different polypeptide backbone, and this structural difference leads to discrete folding options, then we must conclude that the code is truly multi-dimensional and it carries stereochemical information. A different “look” to the above sequence could take on any one of roughly 4000 appearances to us, assuming that we are leucine in E. Coli as described.
This is the view of a simple case, tetra-leucine. What is the view of our hypothetical 100 residue backbone? How many different topologies can the tRNA scaffolds generate? How do these differences translate into the information content of proteins? These are all good questions that to my eye haven’t even been asked, let alone answered. This institutional absence of curiosity is ludicrous, because after all, this is the genetic code. Dogma be damned.
We should realize that every organism can and does have a different complement of tRNA. From this standpoint, the assumption that all peptide bonds emerge from translation in a perfectly identical conformation seems untenable. If translation is causing this lack of homogeneity in peptide bonds then we must identify the information behind this, wherever it may reside.
The first significant impact of this realization is that we can identify the location of the genetic code. The genetic code resides in RNA. The process of translating nucleic acids into amino acids is an entirely RNA phenomenon. It involves many molecules of RNA, not just mRNA, and all involved RNA molecules must be viewed as a part of the genetic code. It is a stereochemical phenomenon, not one-dimensional sequence-only, and it contains stereochemical information. Codons reside on mRNA. They specify tRNA, but each tRNA interacts with every other tRNA. It is the tRNA-tRNA interaction in a sequence that defines every peptide bond in a protein. Finally, rRNA interacts with both mRNA and tRNA to complete the apparatus that builds each and every peptide bond. It is this complement of RNA – mRNA, tRNA, and rRNA – that defines the genetic code of protein synthesis for each organism.
The second significant impact of this thinking is the conclusion that the genetic code is far from universal. Not even the assignment table is universal, but this is just the tip of the non-universal iceberg. There is potentially a universal code, or code limit, but no known organism would need or ever approach this limit. If tRNA is an integral part of the code, and tRNA varies widely between organisms, then the code varies widely between organisms as well. We can visualize this with the following pair of schematics.
This is a return to the visualization of codons matching with the tetrahedrons of amino acids. This is not complete, and therefore it is not an accurate representation of “the genetic code”. This is a relatively complete representation of the correlation table, or assignment table of codons to amino acids, but it ignores many known features of the code. This is a codon table, and codons are all about mRNA. To this we now must add visualization for a network of tRNA.
This is a more accurate picture of what a genetic code might ‘look’ like in a single organism. Change a single nucleic acid in a single tRNA and you have probably changed the code in that organism. This is a truly networked view of the genetic code. It is a dynamic network of RNA.
The really fun thing is that we can now standardize, objectify, and maximally compress this view. That is the view of the Rafiki map. The map itself does not illustrate specific tRNA populations, but it provides the most logical foundation into which they can be placed. The relative positions of all the code’s components can now take shape.

18:48
|
|
This entry was posted on 18:48
You can follow any responses to this entry through
the RSS 2.0 feed.
You can leave a response,
or trackback from your own site.
Subscribe to:
Post Comments (Atom)
0 comments:
Post a Comment